Note
Go to the end to download the full example code.
Relion Projection Interoperability¶
In this tutorial we compare projections generated by Relion
with projections generated by ASPIRE’s Simulation class.
Both sets of projections are generated using a downsampled
volume map of a 70S Ribosome, absent of noise and CTF corruption.
import os
import numpy as np
from aspire.source import RelionSource, Simulation
from aspire.volume import Volume
Load Relion Projections¶
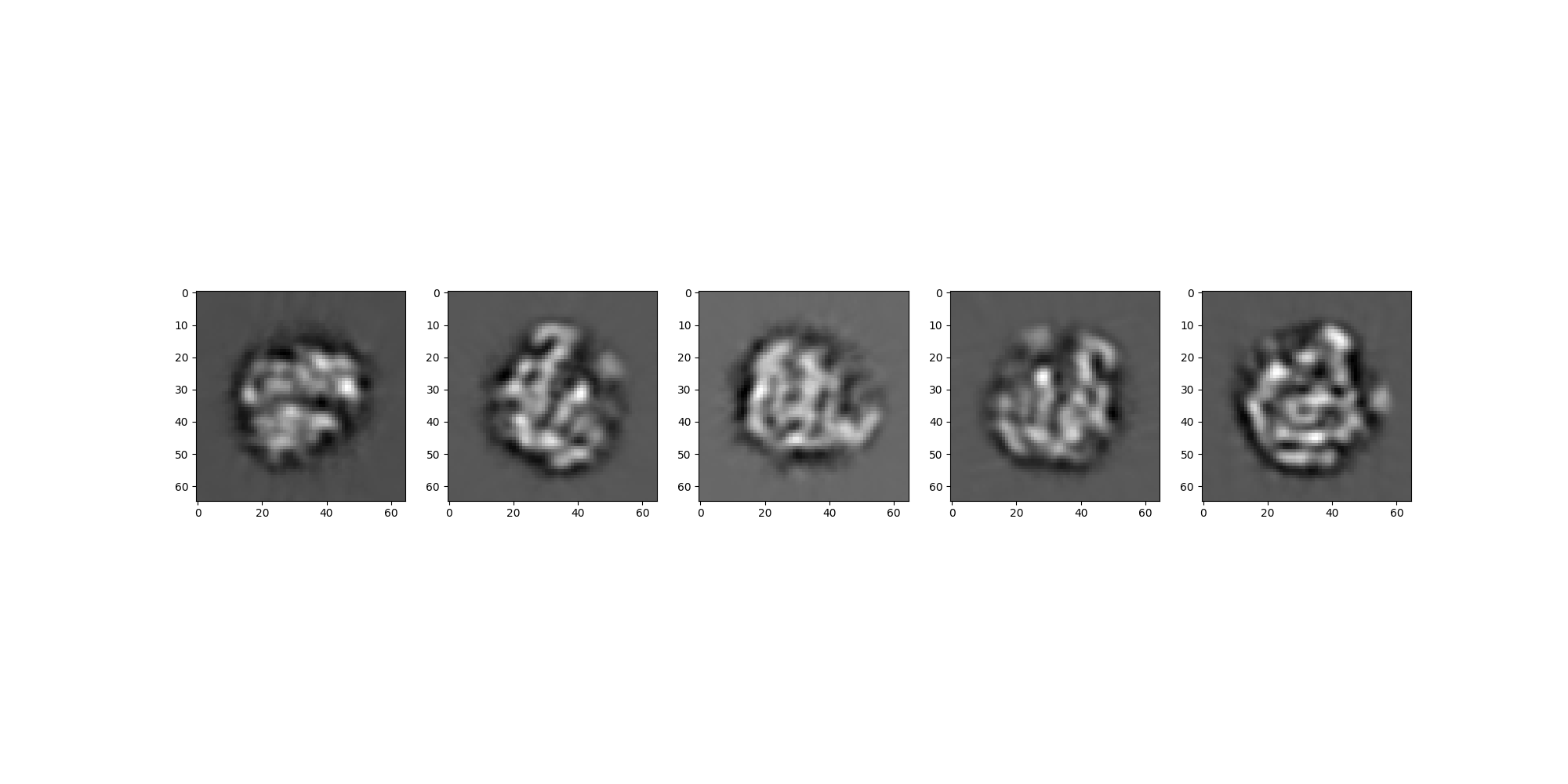
We load the Relion projections as a RelionSource and view the images.
starfile = os.path.join(os.path.dirname(os.getcwd()), "data", "rln_proj_65.star")
rln_src = RelionSource(starfile, pixel_size=1)
rln_src.images[:].show(colorbar=False)
Note
The projections above were generated in Relion using the following command:
relion_project --i clean70SRibosome_vol_65p.mrc --nr_uniform 3000 --angpix 5
For this tutorial we take a subset of these projections consisting of the first 5 images.
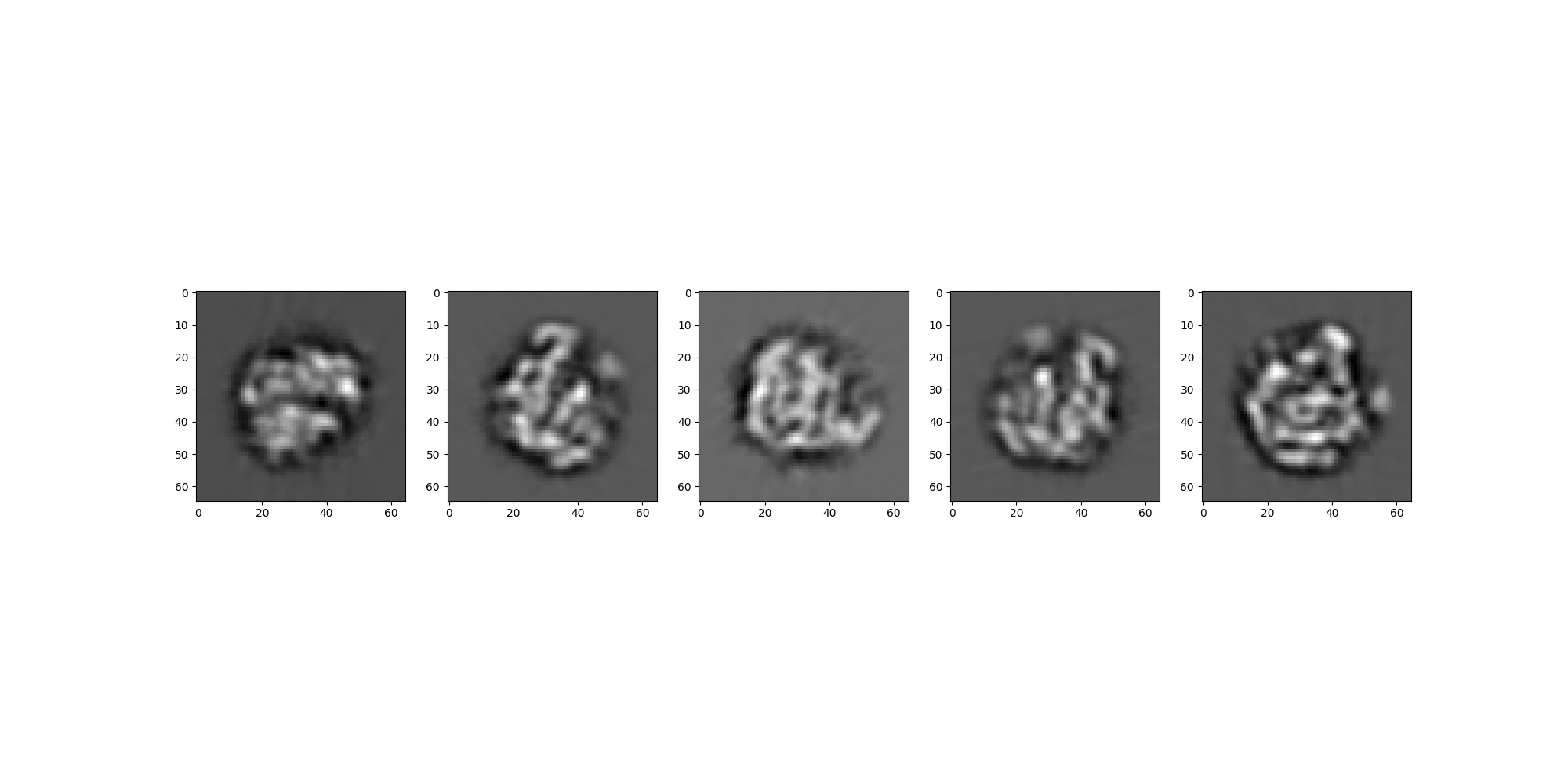
Generate Projections using Simulation¶
Using the metadata associated with the RelionSource and the same volume
we generate an analogous set of projections with ASPIRE’s Simulation class.
# Load the volume from file as a ``Volume`` object.
filepath = os.path.join(
os.path.dirname(os.getcwd()), "data", "clean70SRibosome_vol_65p.mrc"
)
vol = Volume.load(filepath, dtype=rln_src.dtype)
# Create a ``Simulation`` source using metadata from the RelionSource projections.
# Note, for odd resolution Relion projections are shifted from ASPIRE projections
# by 1 pixel in x and y.
sim_src = Simulation(
n=rln_src.n,
vols=vol,
offsets=-np.ones((rln_src.n, 2), dtype=rln_src.dtype),
amplitudes=rln_src.amplitudes,
angles=rln_src.angles,
dtype=rln_src.dtype,
)
sim_src.images[:].show(colorbar=False)
Comparing the Projections¶
We will take a few different approaches to comparing the two sets of projection images.
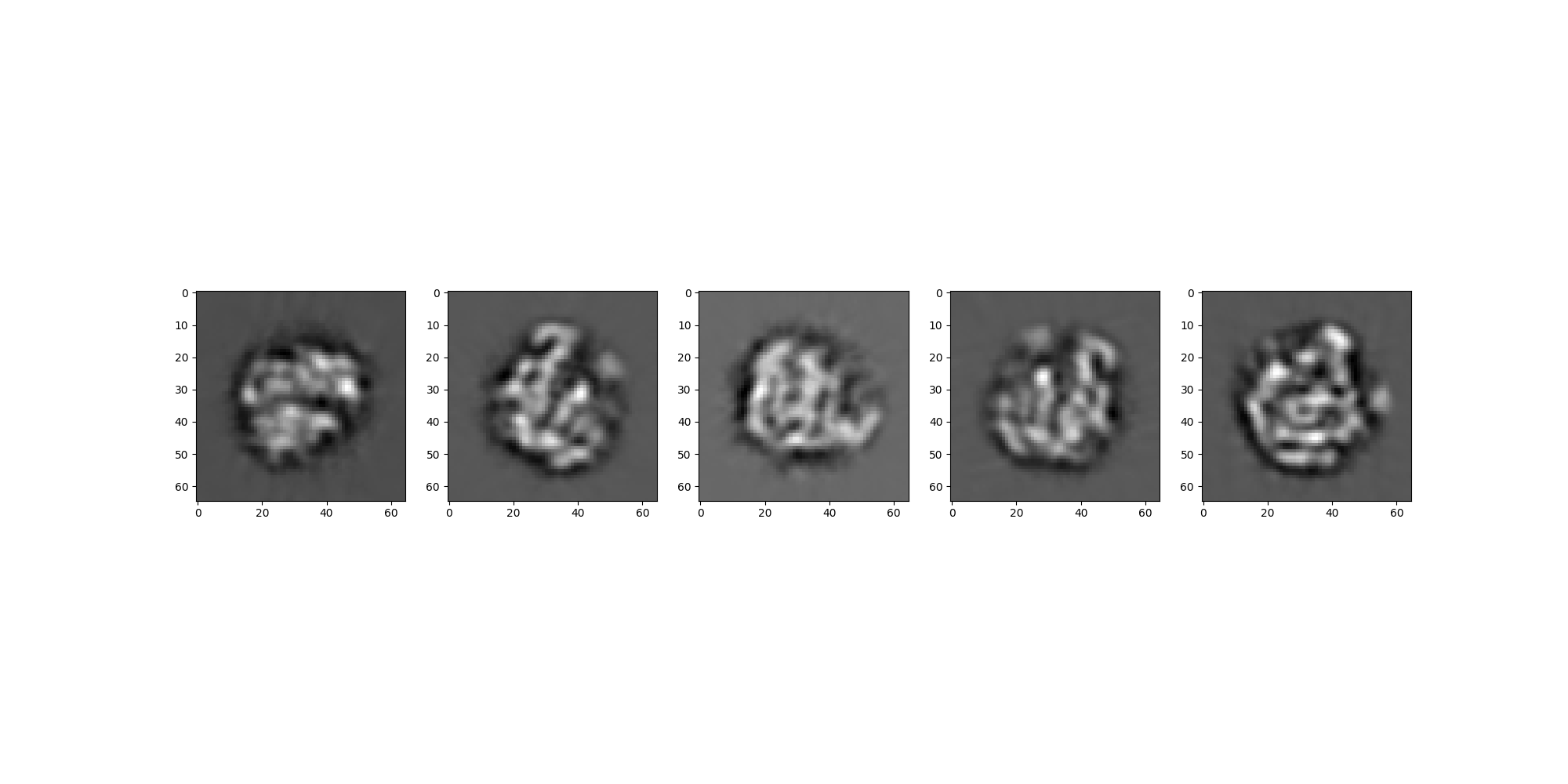
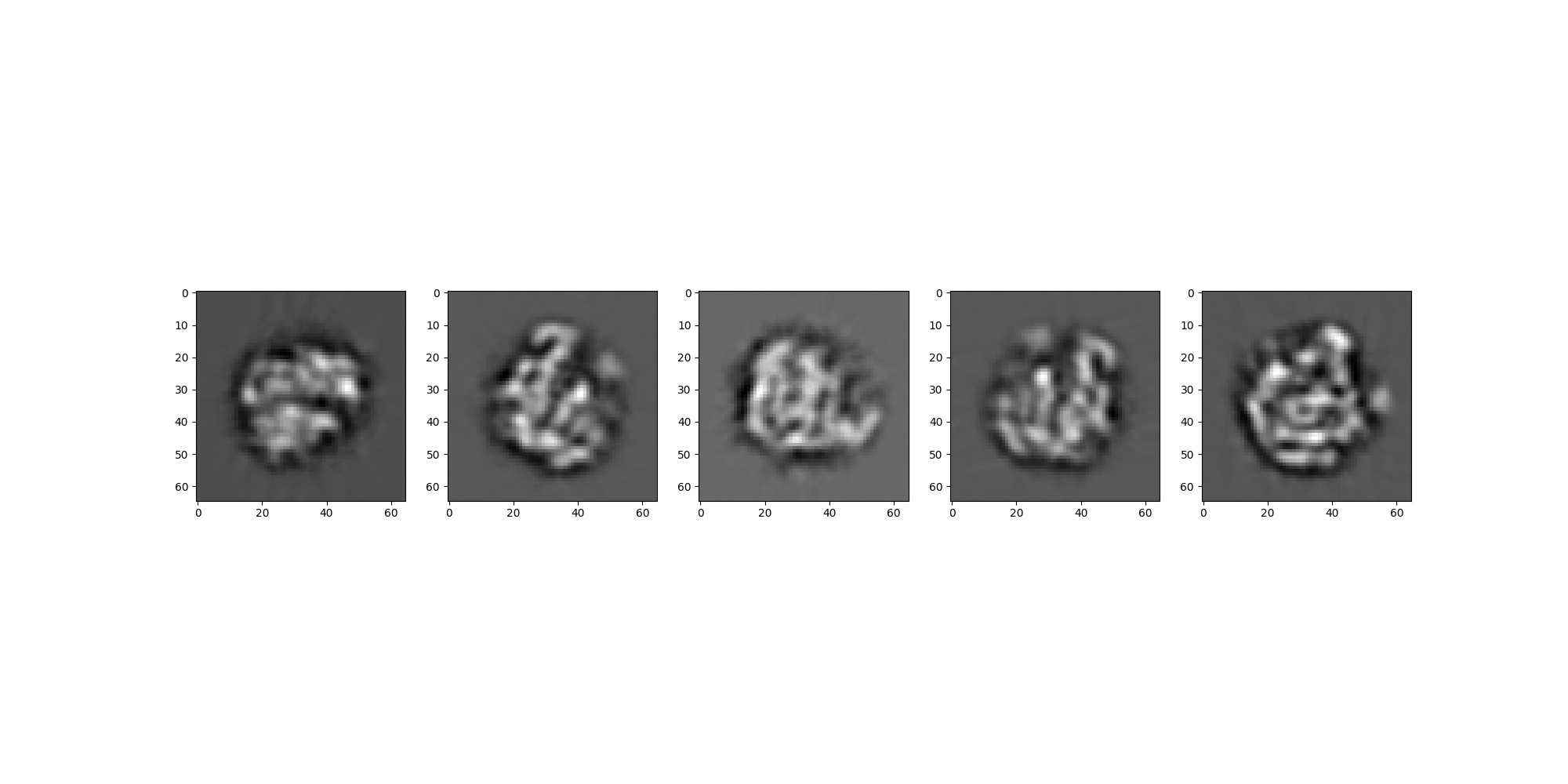
Visual Comparison¶
We’ll first look at a side-by-side of the two sets of images to confirm visually that the projections are taken from the same viewing angles.
rln_src.images[:].show(colorbar=False)
sim_src.images[:].show(colorbar=False)
Fourier Ring Correlation¶
Additionally, we can compare the two sets of images using the FRC. Note that the images are tightly correlated up to a high resolution of 2 pixels.
rln_src.images[:].frc(sim_src.images[:], cutoff=0.143, plot=True)
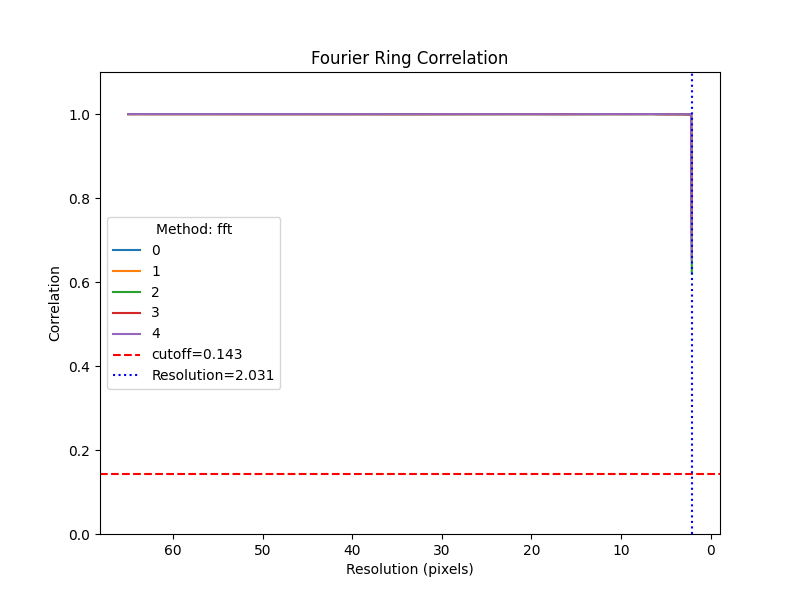
(array([2.03125, 2.03125, 2.03125, 2.03125, 2.03125]), array([[0.99995106, 0.99975914, 0.9997224 , 0.9997412 , 0.9998079 ,
0.99968433, 0.99985576, 0.9998626 , 0.9996016 , 0.9998017 ,
0.99976003, 0.99937594, 0.99953115, 0.99958944, 0.99938124,
0.99957883, 0.999611 , 0.99948025, 0.9995136 , 0.99945575,
0.9994088 , 0.9995097 , 0.9994239 , 0.9993721 , 0.99928445,
0.9991798 , 0.9993925 , 0.9992381 , 0.9991501 , 0.998856 ,
0.9991438 , 0.68903995],
[0.99996096, 0.9996518 , 0.99941385, 0.9996295 , 0.99953634,
0.99973553, 0.99981254, 0.99972916, 0.99975425, 0.9997602 ,
0.9997864 , 0.9996997 , 0.9996596 , 0.99939144, 0.9995733 ,
0.9993533 , 0.99950606, 0.99955046, 0.9992311 , 0.99947745,
0.999399 , 0.9992753 , 0.9993047 , 0.9992677 , 0.99936014,
0.9992539 , 0.9991879 , 0.99928504, 0.9992361 , 0.99918747,
0.9990526 , 0.6563472 ],
[0.99980867, 0.9999284 , 0.9996264 , 0.999767 , 0.99972284,
0.99960023, 0.9996824 , 0.9997461 , 0.9998087 , 0.9996951 ,
0.9998497 , 0.99968654, 0.99962735, 0.99971575, 0.9995882 ,
0.9995541 , 0.99949145, 0.99931914, 0.9991363 , 0.99951273,
0.99946547, 0.9995991 , 0.999417 , 0.99944955, 0.99925214,
0.9989504 , 0.99913013, 0.99941015, 0.99942446, 0.9992472 ,
0.9988977 , 0.61978275],
[0.99984056, 0.9997645 , 0.9996911 , 0.9998639 , 0.9996304 ,
0.9997984 , 0.99977505, 0.99962395, 0.99978775, 0.999842 ,
0.99985904, 0.9996771 , 0.99947655, 0.99927443, 0.9995293 ,
0.999453 , 0.9995277 , 0.9995889 , 0.9993793 , 0.999444 ,
0.9995143 , 0.99963874, 0.9994448 , 0.99924564, 0.99928606,
0.9992858 , 0.9989985 , 0.9992831 , 0.99888486, 0.9988116 ,
0.99889356, 0.7134848 ],
[0.99992067, 0.99977815, 0.9996207 , 0.9996264 , 0.9994458 ,
0.9997416 , 0.9997853 , 0.99961793, 0.9997694 , 0.9998145 ,
0.99977046, 0.99975216, 0.9997678 , 0.9995889 , 0.99926215,
0.9995358 , 0.9994929 , 0.9995174 , 0.9996956 , 0.99941885,
0.9994483 , 0.9993258 , 0.9993586 , 0.99936575, 0.99932224,
0.99923396, 0.9994343 , 0.99917585, 0.9990704 , 0.99915725,
0.9991648 , 0.6540186 ]], dtype=float32))
Relative Error¶
As Relion and ASPIRE differ in methods of generating projections, the pixel intensity of the images may not correspond perfectly. So we begin by first normalizing the two sets of projections. We then check that the relative error with respect to the frobenius norm is less than 3%.
# Work with numpy arrays.
rln_np = rln_src.images[:].asnumpy()
sim_np = sim_src.images[:].asnumpy()
# Normalize images.
rln_np = (rln_np - np.mean(rln_np)) / np.std(rln_np)
sim_np = (sim_np - np.mean(sim_np)) / np.std(sim_np)
# Assert that error is less than 3%.
error = np.linalg.norm(rln_np - sim_np, axis=(1, 2)) / np.linalg.norm(
rln_np, axis=(1, 2)
)
assert all(error < 0.03)
print(f"Relative per-image error: {error}")
Relative per-image error: [0.02434892 0.02776868 0.02810057 0.02241779 0.02729597]
Total running time of the script: (0 minutes 1.047 seconds)