Note
Go to the end to download the full example code.
Micrograph Sources¶
This tutorial will demonstrate how to set up and use ASPIRE’s
MicrographSource classes.
import os
import tempfile
import numpy as np
from aspire.source import ArrayMicrographSource
Overview¶
MicrographSource is an abstract class which provides access to
three distinct subclasses. The first two are
ArrayMicrographSource and DiskMicrographSource which provide
access to array and disk backed micrograph data respectively.
MicrographSimulation takes a volume and generates projection
images which are aggregated into synthetic microgaphs. The following
illustrates an overview of the interfaces, and the tutorial will go
on to demonstrate common operations for each class.
classDiagram
class MicrographSource{
micrograph_count: int
micrograph_size: int
dtype: np.dtype
+asnumpy()
+dtype
+len()
+repr()
+images[]
+micrograph_count
+micrograph_size
+save()
+show()
}
class ArrayMicrographSource{
micrographs: np.ndarray
}
class DiskMicrographSource{
micrographs_path: str, Path, or list
}
class MicrographSimulation{
volume: Volume
micrograph_size: Optional, int
micrograph_count: Optional, int
particles_per_micrograph: Optional, int
particle_amplitudes: Optional, np.ndarray
projection_angles: Optional, np.ndarray
seed: Optional, int
ctf_filters: Optional, list
noise_adder: Optional, NoiseAdder
boundary: Optional, int
interparticle_distance: Optional, int
+boundary
+centers
+ctf_filters
+clean_images[]
+filter_indices
+get_micrograph_index()
+get_particle_index()
+interparticle_distance
+noise_adder
+simulation
+particle_amplitudes
+particle_box_size
+particle_per_micrograph
+projection_angles
+total_particle_count
+volume
}
MicrographSource <|-- ArrayMicrographSource
MicrographSource <|-- DiskMicrographSource
MicrographSource <|-- MicrographSimulation
MicrographSimulation o-- Volume
MicrographSimulation *-- CTFFilter
MicrographSimulation *-- NoiseAdder
Creating an ArrayMicrographSource¶
An ArrayMicrographSource is populated with an array. For this
demonstration, random data will initialize the object,
then this data will be saved off for use in the next example
(which loads data from files).
# Create an (2,512,512) array of data.
# This represents two (512,512) micrographs.
mgs_np = np.random.rand(2, 512, 512)
# Construct the source
src = ArrayMicrographSource(mgs_np)
# Create a tmp dir for saving the data to.
# This just for ensuring the tutorial script is portable,
tmp_dir = tempfile.TemporaryDirectory()
# Save the data as multiple MRC files
# This method returns a file_list,
# which might be useful for loading or other operations.
file_list = src.save(tmp_dir.name)
Creating a DiskMicrographSource¶
A DiskMicrographSource is populated with str or list
representing the location of MRC files.
from aspire.source import DiskMicrographSource
# Load files in directory
src = DiskMicrographSource(tmp_dir.name)
# Load files from a list
src = DiskMicrographSource(file_list)
Creating a Micrograph Simulation¶
A MicrographSimulation is populated with particle projections
from a Volume, so we’ll begin by generating a Volume.
from aspire.source import MicrographSimulation
from aspire.volume import AsymmetricVolume
# Generate one (100,100,100) ``Volume``.
vol = AsymmetricVolume(
L=100,
C=1,
pixel_size=4,
seed=1234,
dtype=np.float32,
).generate()
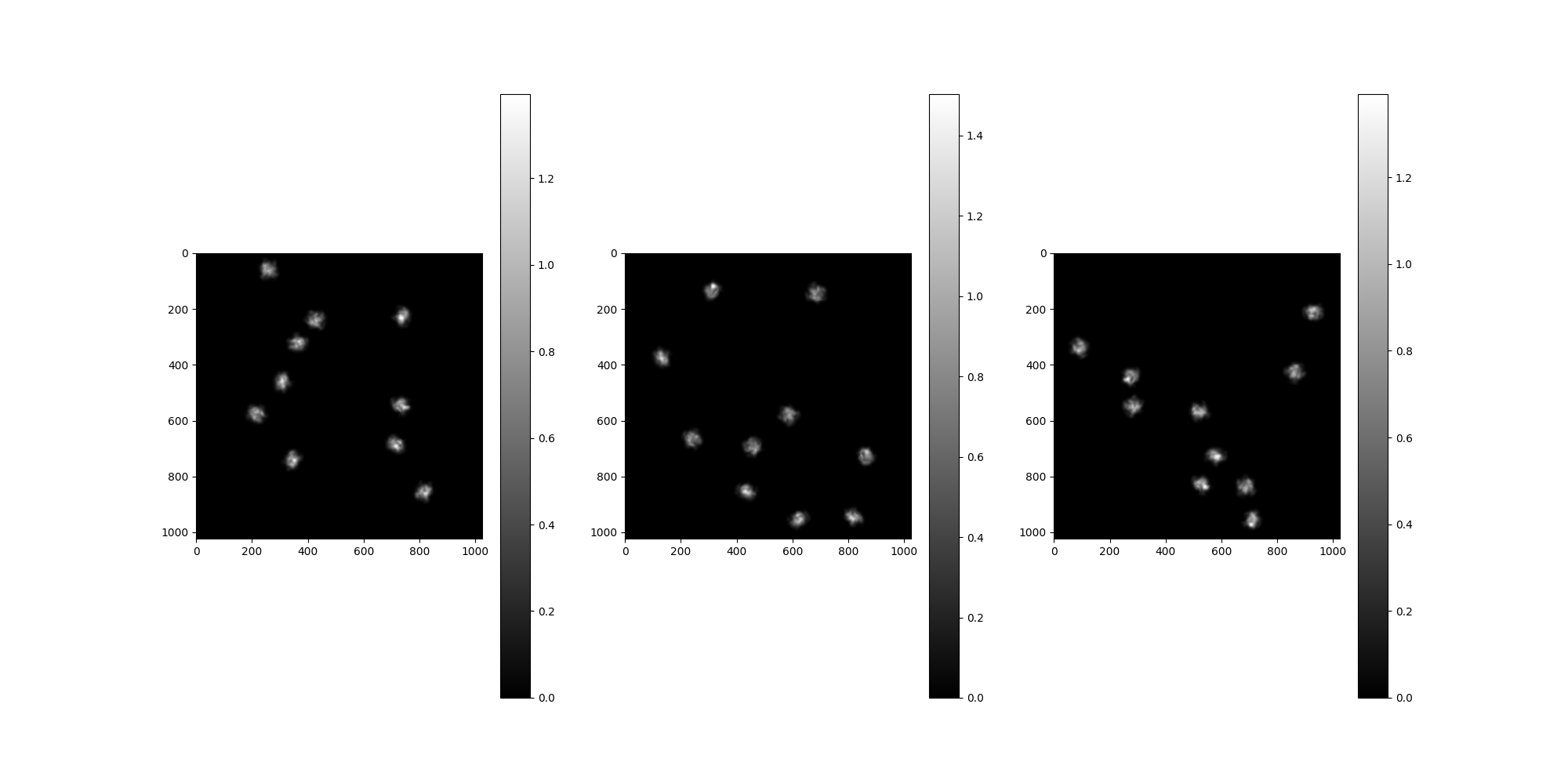
We’ll pass our Volume as an argument and configure our
MicrographSimulation. In this example, the
MicrographSimulation has 4 micrographs of size 1024, each with 10
particles.
n_particles_per_micrograph = 10
n_micrographs = 3
src = MicrographSimulation(
vol,
particles_per_micrograph=n_particles_per_micrograph,
particle_amplitudes=1,
micrograph_size=1024,
micrograph_count=n_micrographs,
seed=1234,
)
# Plot the micrographs
src.images[:].show()
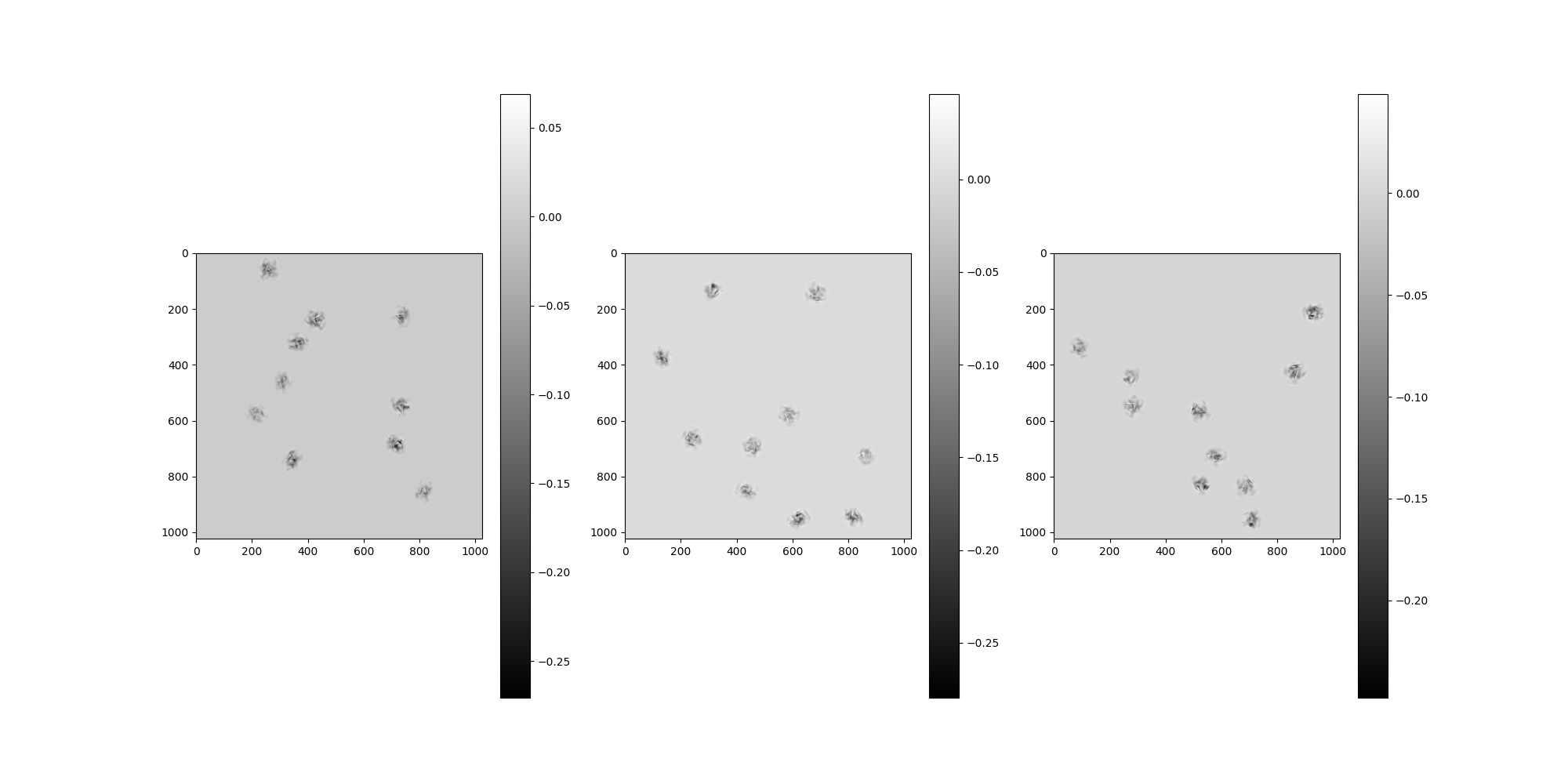
CTF Filters¶
By default, no CTF corruption is configured. To apply CTF filters,
we have to pass them as arguments to the MicrographSimulation.
It is possible to apply a single CTF, different CTF per-micrograph
or different CTF per-particle by configuring a list of matching size.
from aspire.operators import RadialCTFFilter
# Create our CTF Filter and add it to a list.
# This configuration will apply the same CTF to all particles.
ctfs = [
RadialCTFFilter(voltage=200, defocus=15000, Cs=2.26, alpha=0.07, B=0),
]
src = MicrographSimulation(
vol,
particles_per_micrograph=n_particles_per_micrograph,
micrograph_size=1024,
micrograph_count=n_micrographs,
ctf_filters=ctfs,
seed=1234,
)
# Plot the micrographs
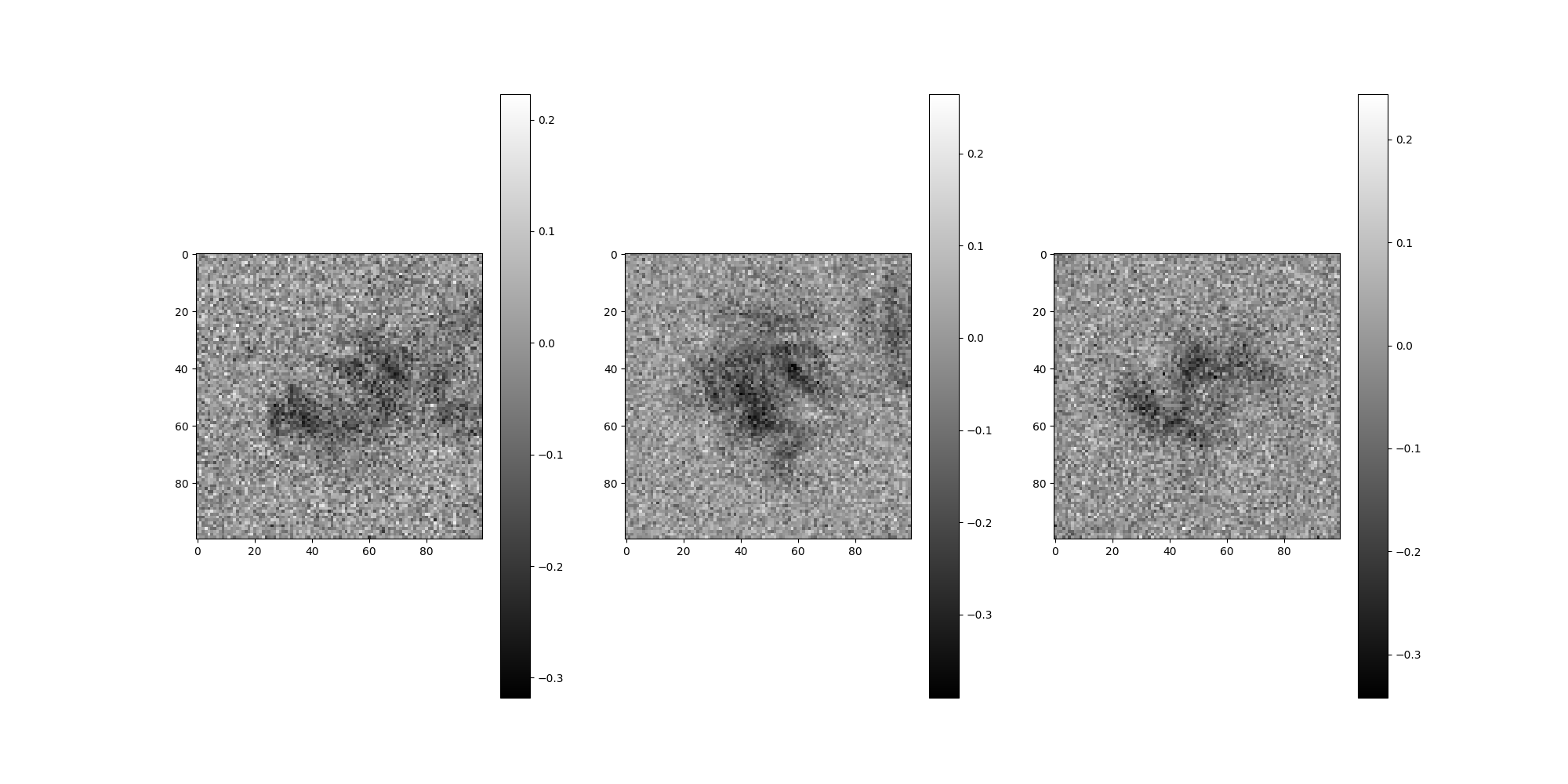
src.images[:].show()
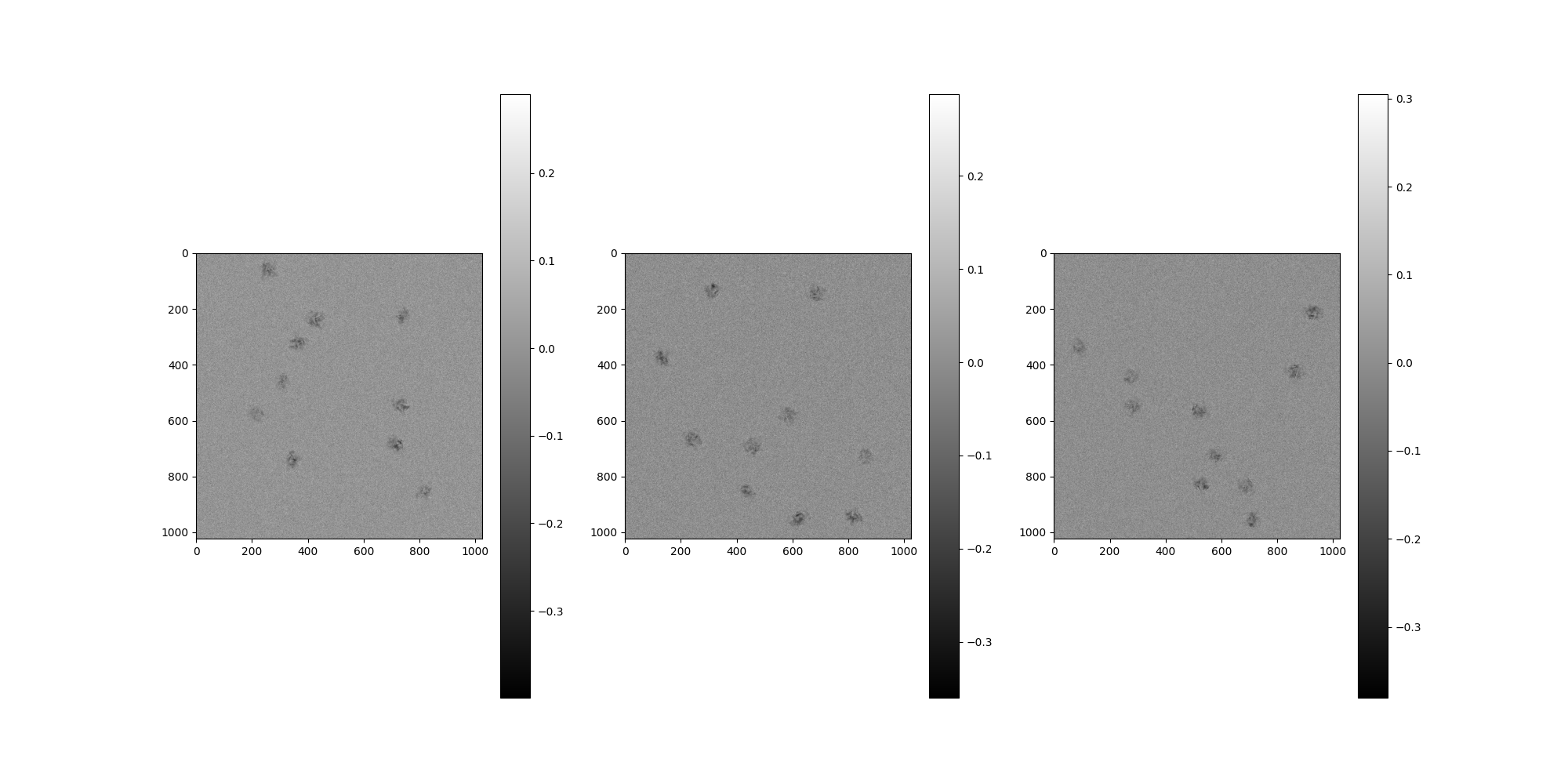
Noise¶
By default, no noise corruption is configured.
To apply noise, pass a NoiseAdder to MicrographSimulation.
from aspire.noise import WhiteNoiseAdder
# Create our noise using WhiteNoiseAdder
noise = WhiteNoiseAdder(4e-3, seed=1234)
# Add noise to our MicrographSimulation using the noise_adder argument
src = MicrographSimulation(
vol,
noise_adder=noise,
particles_per_micrograph=n_particles_per_micrograph,
micrograph_size=1024,
micrograph_count=n_micrographs,
ctf_filters=ctfs,
seed=1234,
)
# Plot the micrographs
src.images[:].show()
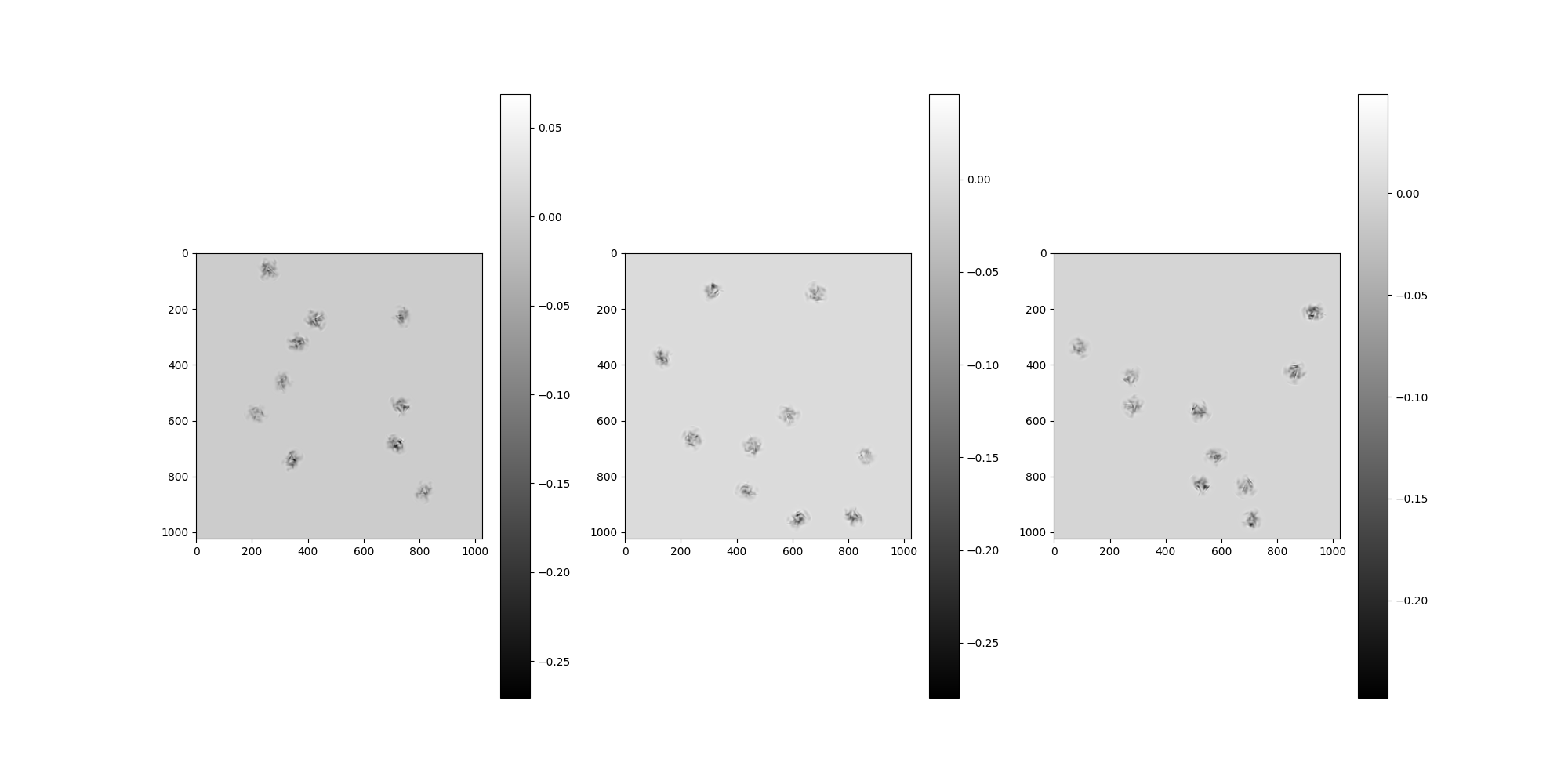
Plot the clean micrographs using the clean_images accessor.
src.clean_images[:].show()
Interparticle Distance¶
By default, particle distance is set to avoid collisions.
We can use the interparticle_distance argument to control the
minimum distance between particle centers.
However, setting this argument too large may generate insufficient centers.
# Let's increase the number of particles to show overlap.
n_particles_per_micrograph = 50
# Set the interparticle distance to 1, which adds at least one pixel
# of separation between center and allows particles to collide.
src = MicrographSimulation(
vol,
interparticle_distance=1,
noise_adder=noise,
particles_per_micrograph=n_particles_per_micrograph,
micrograph_size=1024,
micrograph_count=n_micrographs,
ctf_filters=ctfs,
)
# Plot the micrographs
src.images[:].show()
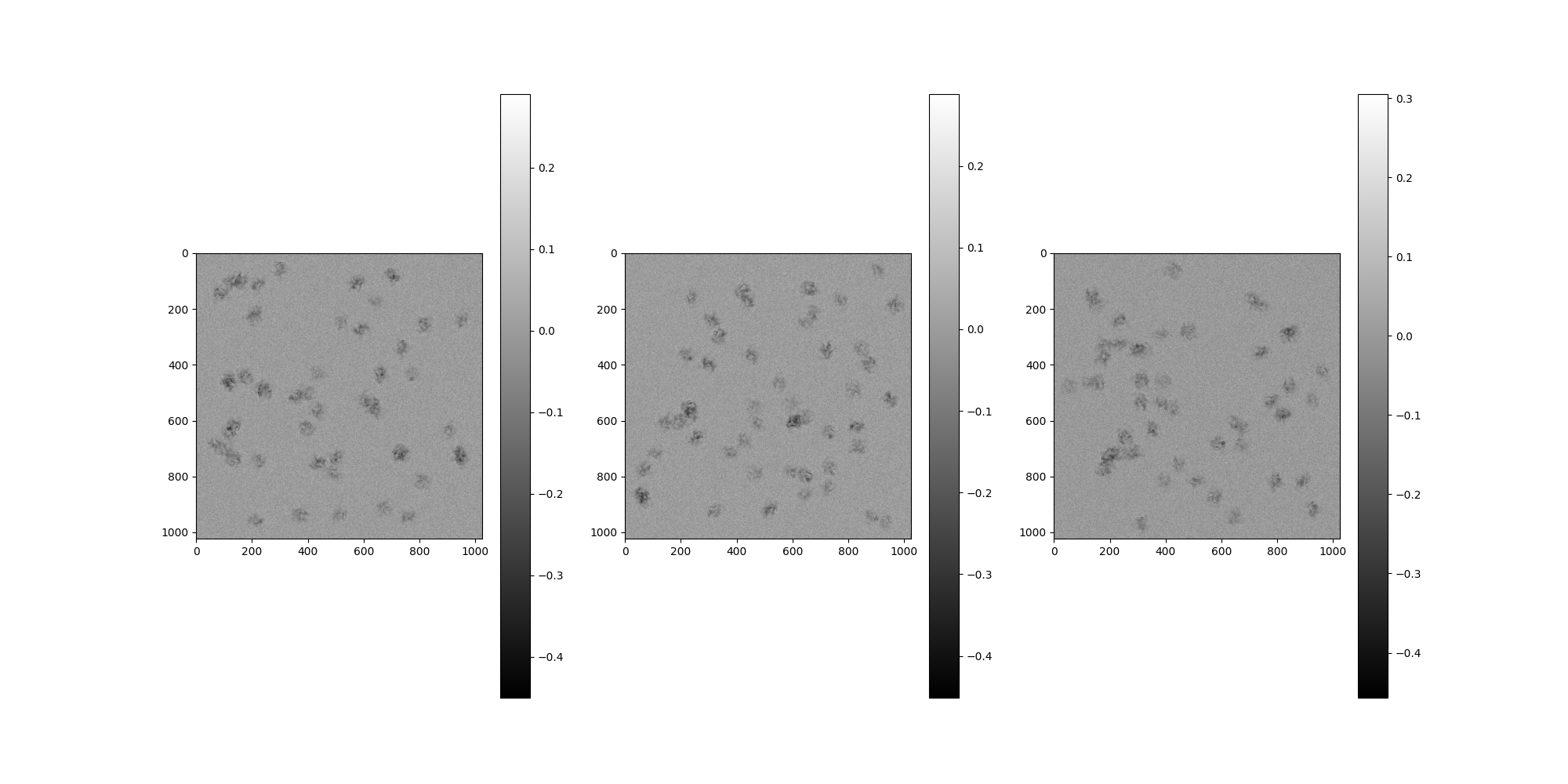
Boundary¶
By default, the boundary is set to half of the particle width,
which will completely contain every particle inside the micrograph.
Setting boundary=0 will allow particles to be placed along the edges.
Positive values (measured in pixels) move the boundaries inward,
while negative values move the boundaries outward.
# Create a micrograph with a negative boundary, allowing particles to
# generate outward.
out_src = MicrographSimulation(
vol,
boundary=-20,
interparticle_distance=1,
noise_adder=noise,
particles_per_micrograph=n_particles_per_micrograph,
micrograph_size=1024,
micrograph_count=n_micrographs,
ctf_filters=ctfs,
)
# Plot the micrographs
out_src.images[:].show()
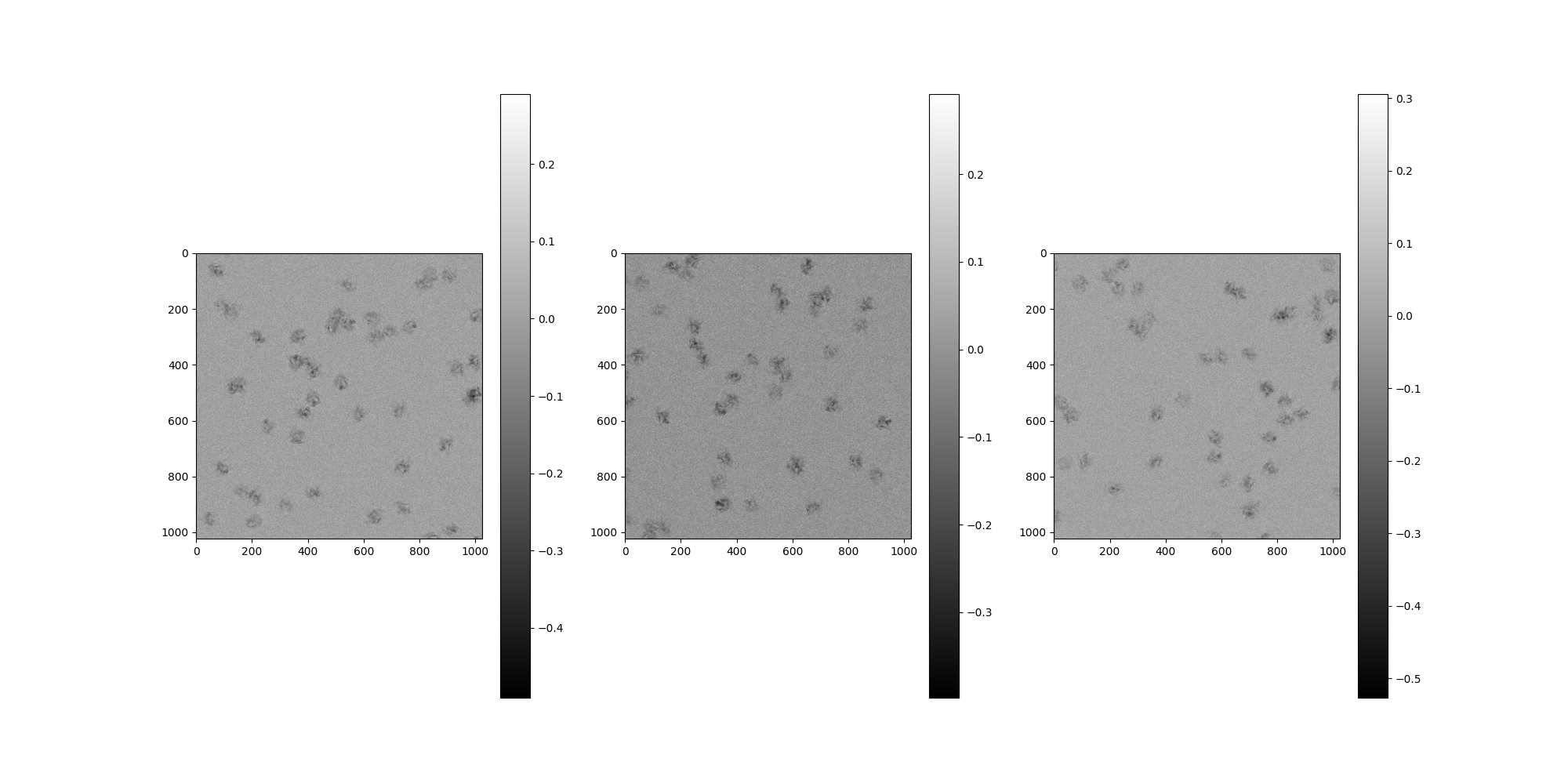
Particle Indices¶
Each particle comes from a Simulation internal to
MicrographSimulation. This simulation can be accessed directly
by the attribute MicrographSimulation.simulation. A map is
provided between each particle’s indexing relative to that
Simulation and micrograph based indexing. This relationship is
demonstrated below.
# Let's choose four random numbers as our global (``Simulation``)
# particle indices from ``test_micrograph=1``.
test_micrograph = 1
n_particles = 3
local_particle_indices = np.random.choice(n_particles_per_micrograph, n_particles)
print(f"Local particle indices: {local_particle_indices}")
Local particle indices: [24 20 36]
We can obtain the individual particle images from our
MicrographSimulation by retrieving their centers and plotting
the boundary boxes.
centers = np.zeros((n_particles, 2), dtype=int)
for i in range(n_particles):
centers[i] = src.centers[test_micrograph][local_particle_indices[i]]
# Let's use the particles' centers and sizes to perform "perfect
# particle picking" on this test micrograph.
p_size = src.particle_box_size
micrograph_picked_particles = np.zeros(
(
n_particles,
src.particle_box_size,
src.particle_box_size,
)
)
for i, center in enumerate(centers):
x, y = center[0], center[1]
# Calculate the square of the particle
particle = src.clean_images[test_micrograph].asnumpy()[0][
x - p_size // 2 : x + p_size // 2, y - p_size // 2 : y + p_size // 2
]
micrograph_picked_particles[i] = particle
# Let's plot and look at the particles!
from aspire.image import Image
Image(micrograph_picked_particles)[:].show()
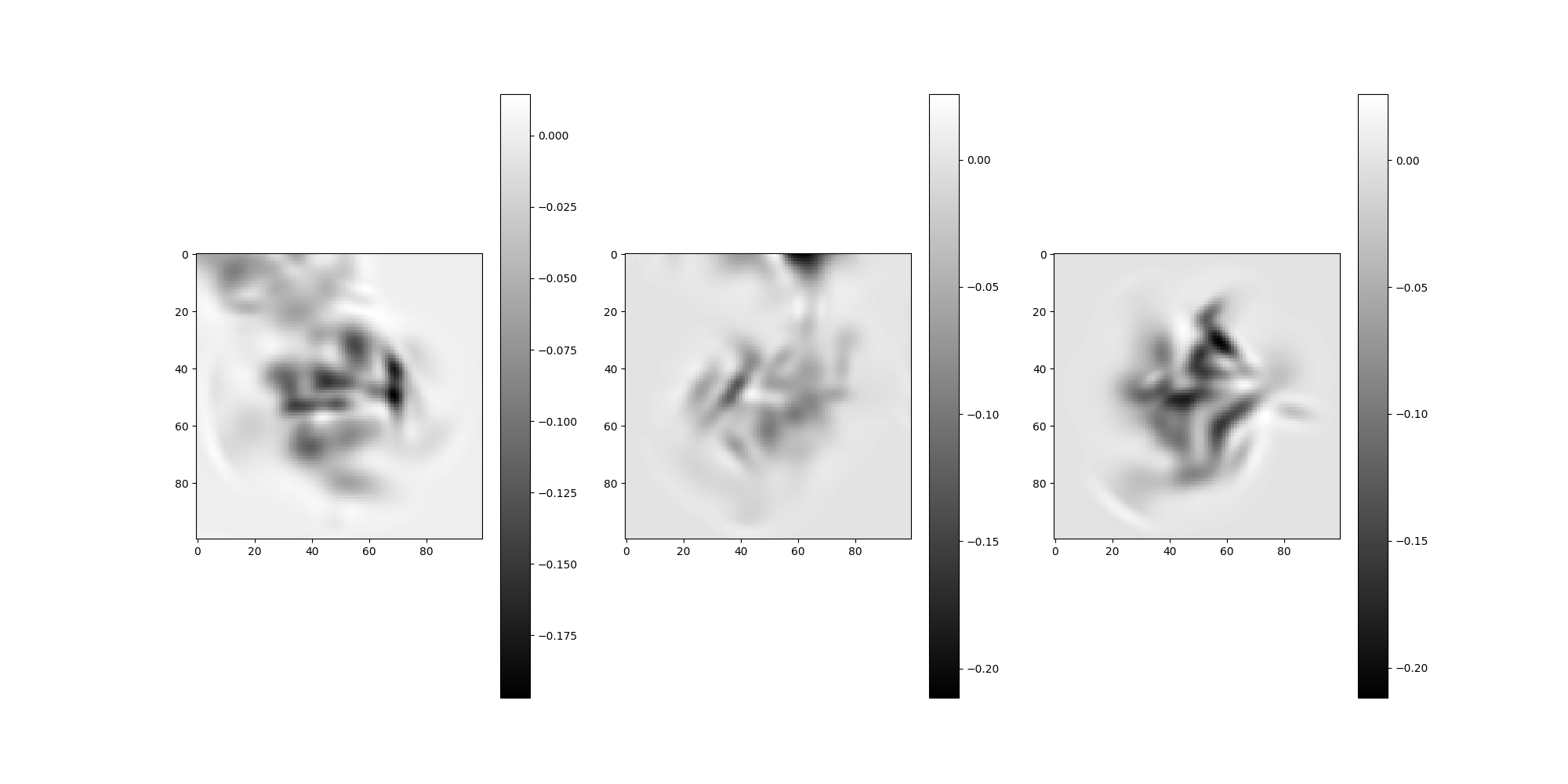
Note
There may be overlap with nearby particles in the above images.
To reduce overlap, increase interparticle_distance.
Let’s find the images from the Simulation using the
get_particle_indices method to retrieve their global indices.
global_particle_indices = np.zeros((n_particles), dtype=int)
for i in range(n_particles):
global_particle_indices[i] = src.get_particle_indices(
test_micrograph, local_particle_indices[i]
)
# Plot the simulation's images
src.simulation.images[global_particle_indices].show()
We can check if these global indices match our local particle
indices with the get_micrograph_index method.
check_local_indices = np.zeros((n_particles), dtype=int)
for i in range(n_particles):
# Get each particle's corresponding micrograph index and local particle index
micrograph_index, check_local_indices[i] = src.get_micrograph_index(
global_particle_indices[i]
)
assert micrograph_index == 1
np.testing.assert_array_equal(local_particle_indices, check_local_indices)
print(f"Local particle indices: {check_local_indices}")
Local particle indices: [24 20 36]
Saving a MicrographSimulation¶
In addition to saving the raw MRC files, MicrographSimulation
populates STAR files with the particle centers, particle box size
(rlnImageSize), and projection rotations. Additionally, CTF
parameters are saved when CTF is used in the simulation. Each
micrograph will have a corresponidng STAR file. The collection of
these files are returned from MicrographSimulation.save as a
list of tuples which is designed to work directly with
CentersCoordinateSource.
from aspire.source import CentersCoordinateSource
# Save the simulation
results = src.save(os.path.join(tmp_dir.name, "mg_sim"))
# Review the resulting files
print(results)
[('/tmp/tmpu21t6i2l/mg_sim/micrograph_0.mrc', '/tmp/tmpu21t6i2l/mg_sim/micrograph_0.star'), ('/tmp/tmpu21t6i2l/mg_sim/micrograph_1.mrc', '/tmp/tmpu21t6i2l/mg_sim/micrograph_1.star'), ('/tmp/tmpu21t6i2l/mg_sim/micrograph_2.mrc', '/tmp/tmpu21t6i2l/mg_sim/micrograph_2.star')]
# Review the example STAR file contents
with open(results[0][1], "r") as f:
print(f.read())
data_
loop_
_rlnImageName
_rlnImageSize
_rlnCoordinateX
_rlnCoordinateY
_rlnAngleRot
_rlnAngleTilt
_rlnAnglePsi
_rlnVoltage
_rlnDefocusU
_rlnDefocusV
_rlnDefocusAngle
_rlnSphericalAberration
_rlnAmplitudeContrast
000001@micrograph_0 100 357 514 -134.82674 70.42997 85.89395 200.0 15000.0 15000.0 0.0 2.26 0.07
000002@micrograph_0 100 118 467 67.95413 99.76551 37.444767 200.0 15000.0 15000.0 0.0 2.26 0.07
000003@micrograph_0 100 590 277 7.2576485 140.38687 -59.773598 200.0 15000.0 15000.0 0.0 2.26 0.07
000004@micrograph_0 100 522 248 63.480827 84.10157 7.4896197 200.0 15000.0 15000.0 0.0 2.26 0.07
000005@micrograph_0 100 223 745 -130.84204 21.15329 -162.84654 200.0 15000.0 15000.0 0.0 2.26 0.07
000006@micrograph_0 100 174 445 -99.030975 149.42964 175.6943 200.0 15000.0 15000.0 0.0 2.26 0.07
000007@micrograph_0 100 299 59 47.95209 136.74622 -166.17136 200.0 15000.0 15000.0 0.0 2.26 0.07
000008@micrograph_0 100 659 434 91.8869 100.16265 -157.42801 200.0 15000.0 15000.0 0.0 2.26 0.07
000009@micrograph_0 100 117 647 -30.354961 75.08341 13.788602 200.0 15000.0 15000.0 0.0 2.26 0.07
000010@micrograph_0 100 735 339 46.58517 129.15901 -172.69316 200.0 15000.0 15000.0 0.0 2.26 0.07
000011@micrograph_0 100 491 789 110.339386 99.2707 85.04628 200.0 15000.0 15000.0 0.0 2.26 0.07
000012@micrograph_0 100 809 818 -23.406391 52.53211 -139.94836 200.0 15000.0 15000.0 0.0 2.26 0.07
000013@micrograph_0 100 436 431 134.82375 125.791466 127.273315 200.0 15000.0 15000.0 0.0 2.26 0.07
000014@micrograph_0 100 212 212 88.836 131.61661 170.09749 200.0 15000.0 15000.0 0.0 2.26 0.07
000015@micrograph_0 100 374 937 71.1484 50.87036 117.835976 200.0 15000.0 15000.0 0.0 2.26 0.07
000016@micrograph_0 100 159 101 63.209724 61.319878 -144.78423 200.0 15000.0 15000.0 0.0 2.26 0.07
000017@micrograph_0 100 727 717 171.41907 103.669914 162.73087 200.0 15000.0 15000.0 0.0 2.26 0.07
000018@micrograph_0 100 434 566 148.2073 23.667522 -167.71777 200.0 15000.0 15000.0 0.0 2.26 0.07
000019@micrograph_0 100 514 940 124.225586 101.704155 -7.356131 200.0 15000.0 15000.0 0.0 2.26 0.07
000020@micrograph_0 100 644 172 -117.36976 54.44209 61.728706 200.0 15000.0 15000.0 0.0 2.26 0.07
000021@micrograph_0 100 758 943 -162.91643 43.91972 100.298325 200.0 15000.0 15000.0 0.0 2.26 0.07
000022@micrograph_0 100 394 628 -0.5854195 75.2847 -139.51906 200.0 15000.0 15000.0 0.0 2.26 0.07
000023@micrograph_0 100 87 144 171.61751 15.32889 111.023254 200.0 15000.0 15000.0 0.0 2.26 0.07
000024@micrograph_0 100 204 235 -33.304127 57.931633 -88.27734 200.0 15000.0 15000.0 0.0 2.26 0.07
000025@micrograph_0 100 775 436 24.332895 108.68852 9.214272 200.0 15000.0 15000.0 0.0 2.26 0.07
000026@micrograph_0 100 702 85 166.2759 55.460697 -116.08049 200.0 15000.0 15000.0 0.0 2.26 0.07
000027@micrograph_0 100 816 257 -17.669033 164.46213 154.5533 200.0 15000.0 15000.0 0.0 2.26 0.07
000028@micrograph_0 100 245 494 -154.18813 160.12997 93.87625 200.0 15000.0 15000.0 0.0 2.26 0.07
000029@micrograph_0 100 222 116 39.215176 119.0958 -38.1352 200.0 15000.0 15000.0 0.0 2.26 0.07
000030@micrograph_0 100 499 733 108.81932 61.365723 -178.08334 200.0 15000.0 15000.0 0.0 2.26 0.07
000031@micrograph_0 100 947 736 109.85279 42.58259 -149.33972 200.0 15000.0 15000.0 0.0 2.26 0.07
000032@micrograph_0 100 134 621 36.03223 105.68081 -35.86676 200.0 15000.0 15000.0 0.0 2.26 0.07
000033@micrograph_0 100 106 707 -47.898327 33.21698 55.839603 200.0 15000.0 15000.0 0.0 2.26 0.07
000034@micrograph_0 100 233 487 -23.364729 81.33271 -88.90829 200.0 15000.0 15000.0 0.0 2.26 0.07
000035@micrograph_0 100 576 110 79.40814 74.74897 -22.989313 200.0 15000.0 15000.0 0.0 2.26 0.07
000036@micrograph_0 100 439 755 -92.107895 65.83309 -104.68323 200.0 15000.0 15000.0 0.0 2.26 0.07
000037@micrograph_0 100 395 504 -155.39862 136.76486 -178.96898 200.0 15000.0 15000.0 0.0 2.26 0.07
000038@micrograph_0 100 738 721 47.220955 58.597233 144.94357 200.0 15000.0 15000.0 0.0 2.26 0.07
000039@micrograph_0 100 952 243 78.088165 78.69344 2.2200243 200.0 15000.0 15000.0 0.0 2.26 0.07
000040@micrograph_0 100 631 543 -139.73576 161.4182 -51.54953 200.0 15000.0 15000.0 0.0 2.26 0.07
000041@micrograph_0 100 903 635 -42.63034 141.55984 150.86116 200.0 15000.0 15000.0 0.0 2.26 0.07
000042@micrograph_0 100 599 527 -83.30669 48.701714 132.33043 200.0 15000.0 15000.0 0.0 2.26 0.07
000043@micrograph_0 100 215 962 -171.20354 59.49243 -103.206924 200.0 15000.0 15000.0 0.0 2.26 0.07
000044@micrograph_0 100 120 455 -85.77694 16.362684 58.064697 200.0 15000.0 15000.0 0.0 2.26 0.07
000045@micrograph_0 100 135 738 -176.21182 115.328545 -104.11368 200.0 15000.0 15000.0 0.0 2.26 0.07
000046@micrograph_0 100 937 717 -3.172558 117.60184 176.06554 200.0 15000.0 15000.0 0.0 2.26 0.07
000047@micrograph_0 100 71 684 -102.14765 63.155346 49.60995 200.0 15000.0 15000.0 0.0 2.26 0.07
000048@micrograph_0 100 644 568 130.81349 123.28017 78.281044 200.0 15000.0 15000.0 0.0 2.26 0.07
000049@micrograph_0 100 672 915 146.05128 137.0732 -30.214369 200.0 15000.0 15000.0 0.0 2.26 0.07
000050@micrograph_0 100 130 103 86.410545 123.06443 121.30503 200.0 15000.0 15000.0 0.0 2.26 0.07
img_src = CentersCoordinateSource(
results,
src.pixel_size,
src.particle_box_size,
)
# Show the first five images from the image source.
img_src.images[:3].show()
# Cleanup the tmp_dir
tmp_dir.cleanup()
Total running time of the script: (0 minutes 14.773 seconds)